Figure 1. C2inform assay protocol
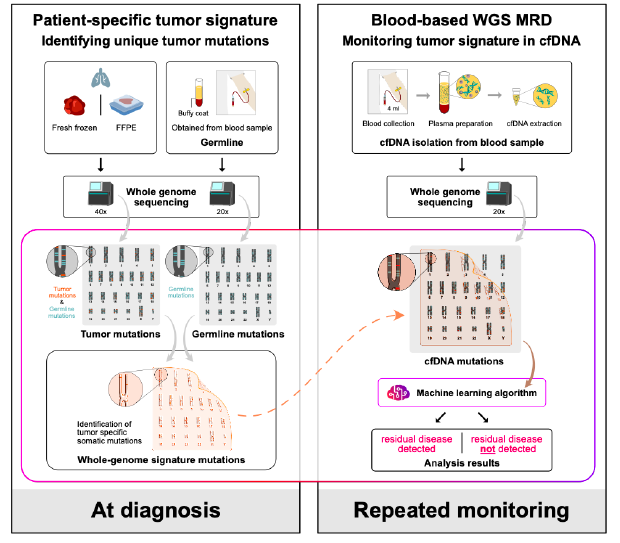
Figure 2. Study design
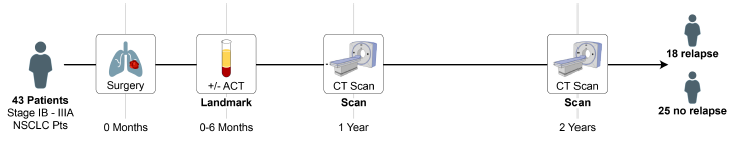
Table 1. Patient Characteristics
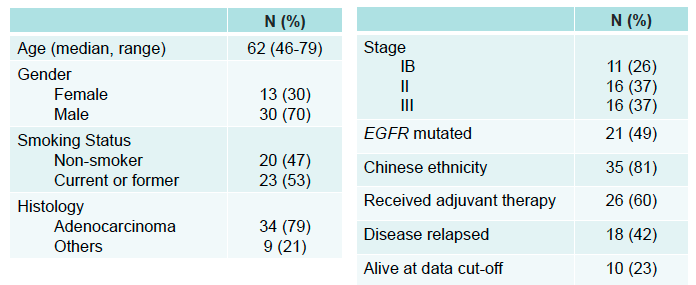
Figure 3. The association of relapse with presence of ctDNA in (A) the landmark cohort and (B) the EGFR mutated and wild type (WT) subgroups
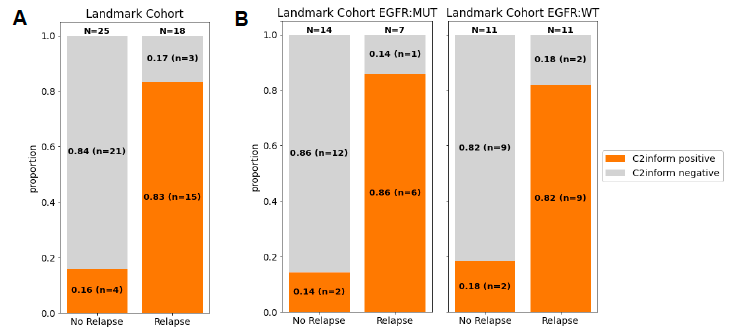
ctDNA was detected (C2inform positive) in 83% of patients that relapsed (sensitivity 83%), compared to 16% that did not relapse (specificity 84%)
Figure 4. Plasma samples from 43 patients in the landmark cohort were collected post-surgery and analyzed for the presence of ctDNA (C2inform positive). All post-surgery plasma samples are shown.
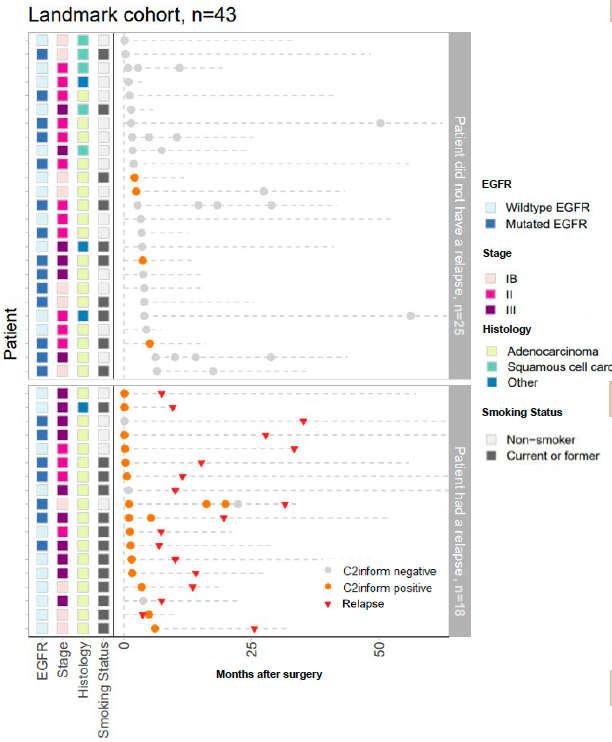
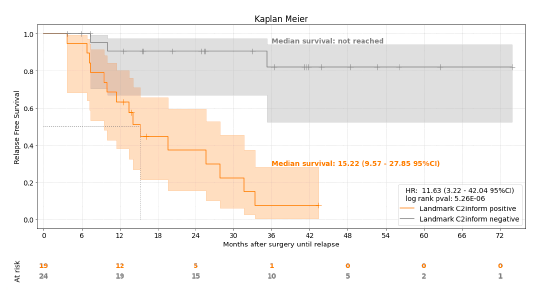
Figure 5. Association of C2inform status at landmark with relapse

Overview: The pivotal role of early detection of cancer recurrence and the precise monitoring of minimal residual disease (MRD) after surgery cannot be overstated, especially in the context of non-small cell lung cancer (NSCLC) where circulating tumor DNA (ctDNA) plays a critical role in clinical outcomes. To address the challenges of ctDNA detection in NSCLC patients undergoing potentially curative resections, we adopted a cutting-edge whole genome sequencing (WGS) method, MRDetect, designed for unmatched sensitivity and specificity in ctDNA analysis.
Study Design: Our exploratory study aimed to assess the efficacy of the MRDetect method across several key stages: before surgery, immediately after, and during subsequent follow-ups, in patients diagnosed with stage IB-IIIA NSCLC. Participants were monitored through standard computed tomography imaging, while ctDNA was meticulously extracted from plasma samples. The MRDetect technique, leveraging WGS with a tumor-specific approach and enhanced by AI-driven error correction models, was pivotal in distinguishing ctDNA signals amidst a backdrop of low tumor burden, thereby providing a nuanced prognosis for recurrence risk. This study also evaluated the assay’s predictive power for recurrence, based on critical samples collected at a median of 1.6 months post-operation.
Outcomes: The pilot encompassed 52 NSCLC patients, yielding 88 plasma samples over a median follow-up period of 32.6 months. Notably, among 43 patients with critical post-operative samples, the median age was 62, with a predominant male demographic (70%) and a high prevalence of adenocarcinoma (79%) and EGFR mutations (49%). The MRDetect assay demonstrated a commendable sensitivity of 83% in detecting radiological recurrence, including a remarkable 86% sensitivity in patients with EGFR mutations. The median relapse-free survival for patients identified as MRD positive by MRDetect was 15.2 months. Moreover, the assay showcased an 84% specificity in patients without recurrence, with consistent performance across EGFR mutation statuses. Longitudinal analysis further confirmed the assay’s high specificity (93%) in predicting the absence of recurrence.
Conclusion: The MRDetect platform, integrating WGS with sophisticated AI algorithms, proves to be a potent tool in identifying MRD with high accuracy in NSCLC patients, irrespective of EGFR mutation status. As the landscape of adjuvant therapies for NSCLC evolves, possessing such an ultra-sensitive MRD assay is invaluable for personalizing treatment plans, optimizing the selection and timing of adjuvant therapies to enhance patient outcomes.
