Figure 1. Schematic of Clinical Sample Collection

Molecular and data analysis Protocol
- Genomic DNA from tumor/germline pairs (n=67) and cfDNA from 510 plasma samples were isolated.
- WGS of tumor/germline pairs followed by genome-wide integration of somatic mutations, enriched by signal processing and AI-based error suppression, were used to generate patient-specific tumor signature (Figure 2).
- Patient-specific somatic variants were used for detecting ctDNA levels in about 15 ng cfDNA from plasma WGS data by the MRDetect5 algorithm (Figure 2)
- Clinical data, such as pathological evaluation and radiographic imaging, were first unblinded after analysis and compared directly to the MRDetect5 plasma call results.
Figure 2. Schematic of Workflow

Figure 3. Concordance between independent laboratories - United States vs. Denmark

Figure 4. Prognostic value of circulating tumor DNA (ctDNA)

Kaplan-Meier survival analysis of recurrence-free survival (RFS)(A+B) and overall survival (OS) (C) stratified by ctDNA status before chemotherapy (or at diagnosis), before cystectomy, and any time after cystectomy.
(A) The follow-up time set to one year after last plasma sample fore each patient if prior to relapse, last CT scan or death.
(B+C) shows Kaplan-Meier analysis for the maximum follow-up time where patients without events were censored at end of follow-up.
Figure 5. Disease courses and longitudinal ctDNA results for the 67 patient study cohort
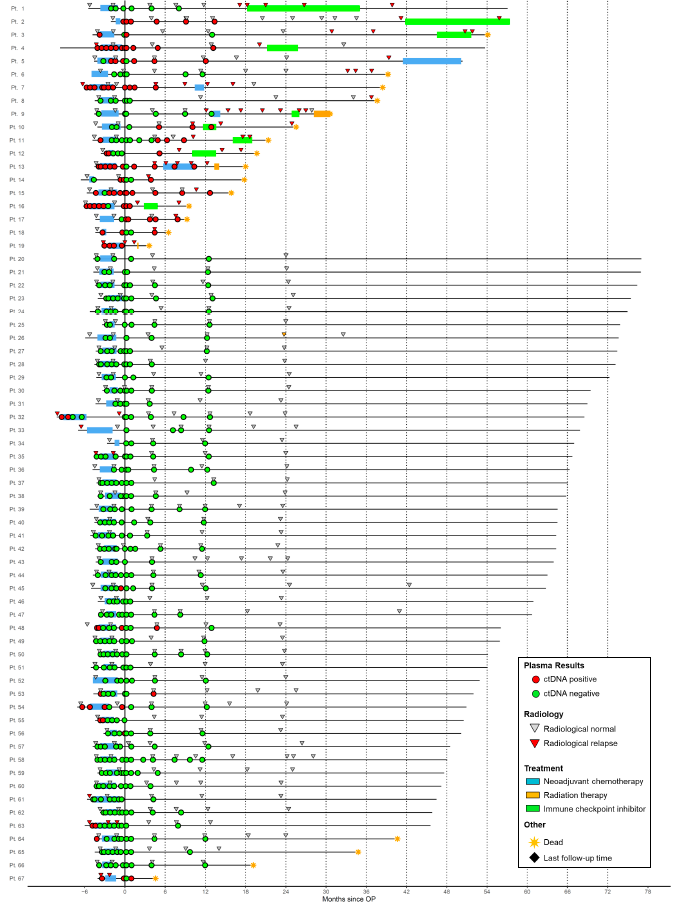
Longitudinal representation of ctDNA results and clinical parameters. Circles represent ctDNA status (see color code in the right box). Treatment and imaging information is indicated for each patient. Patients are separated into two groups on the basis of clinical relapse status. Metastatic relapse (Patient 1-19); Non Relapse (20-67).
Figure 6. Pathological downstaging vs. ctDNA status at diagnosis, pre-CX and post CX

Association between pathological downstaging and ctDNA status before chemotherapy, before cystectomy, and any time after cystectomy.
Figure 7. Prognostic value of circulating tumor DNA (ctDNA) detection - dichotomized analysis

Association between disease recurrence and ctDNA status before chemotherapy, before cystectomy, and any time after cystectomy.
Figure 8. Monitoring tumor fraction during patient disease courses

Figure 9. ctDNA dynamic during NAC as predictive markers of chemotherapy response
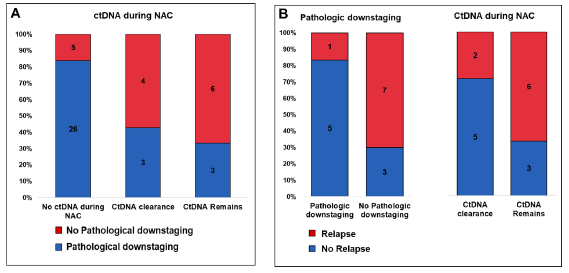
- (A) Association between ctDNA dynamic during NAC and pathological downstaging for ctDNA-negative patients (before and after NAC), patients where ctDNA was cleared during NAC, and patients in whom ctDNA remained
- (B) Association between recurrence and pathological downstaging (left) and ctDNA dynamics during NAC (right) for patients ctDNA-positive before NAC.
Figure 10. Lead time inferred from ctDNA-based recurrence detection

Lead time in days shown as the differences between first molecular relapse (ctDNA positive) and clinical recurrence (radiographic imaging positive). CT Scan corrected Lead time is calculated as the difference between the first CT scan after molecular relapse and clinical recurrence (see box in figure). The CT Scan corrected Lead time is more restrictive and included to provide a fair comparison to radiographic imaging.
Table 1. Patient Characteristics and Demographics (n=67)


Introduction: The integration of neoadjuvant chemotherapy (NAC) followed by radical cystectomy (CX) marks a pivotal advancement in treating localized muscle-invasive bladder cancer (MIBC). Despite these efforts, approximately 45% of MIBC patients experience metastatic recurrence within two years post-CX. The effectiveness of current chemotherapy and immune checkpoint inhibitors (ICI) remains limited, highlighting an urgent need for reliable biomarker tests to track therapeutic responses. Additionally, there’s a growing necessity for biomarkers capable of detecting minimal residual disease (MRD) post-CX to commence timely treatment. Recent studies have revealed that analyzing mutations in cell-free DNA (cfDNA) from blood samples offers a promising avenue for monitoring MRD, yet challenges persist due to the low tumor fraction post-surgery and the minimal sample volume typically obtained from plasma.
Study Design: In our groundbreaking study, we enrolled 140 patients battling MIBC who underwent NAC and CX, divided into an initial test group of 19 patients and a subsequent validation group of 121. We collected and analyzed cfDNA from approximately 1mL of plasma, gathered across various stages: during NAC (to measure response), before cystectomy (to measure response), after surgery (for relapse monitoring), and throughout immunotherapy (during ICI treatment). Our approach utilized whole-genome sequencing (WGS) on tumor/germline pairings and plasma cfDNA, enabling comprehensive genomic alteration detection and precise ctDNA quantification via the MRDetect method.
Findings: Our innovative, personalized WGS model, which merges genome-wide mutation data and copy number variations with sophisticated signal processing and AI-driven error minimization, has set a new standard in precision medicine. By identifying patient-specific somatic variants, we achieved remarkable sensitivity in ctDNA detection from low-input blood samples through WGS. This method successfully identified tumor fractions as low as 8*10^-5. Impressively, in our test cohort, we observed ctDNA post-CX in 7 out of 8 patients who later showed clinical relapse (demonstrating 88% sensitivity) and found no ctDNA in 11 out of 11 patients without clinical relapse (showing 100% specificity). Our method also predicted MRD-based recurrence with a significant lead-time advantage of 9 months over traditional CT imaging. These promising results will be further elaborated at the upcoming AACR 2022 conference.
Conclusion: The evolution of precision oncology necessitates the development of quantitative, non-invasive techniques to customize treatment plans and monitor patient progress closely. Our findings underscore the clinical value of integrating personalized genomic mutations for ultra-sensitive, non-invasive MRD detection and treatment response monitoring, offering a new direction in the clinical management of bladder cancer patients.
