Case Study : 6 y.o. M, Dx of medulloblastoma, Group 4, s/p gross total resection of the cerebral tumor, with spinal leptomeningeal disease. CSF: cytology and ctDNA negative on follow-up.

Case Study : 6 y.o. M, Dx of medulloblastoma, Group 4, s/p gross total resection of the cerebral tumor, with spinal leptomeningeal disease. CSF: cytology and ctDNA negative on follow-up.
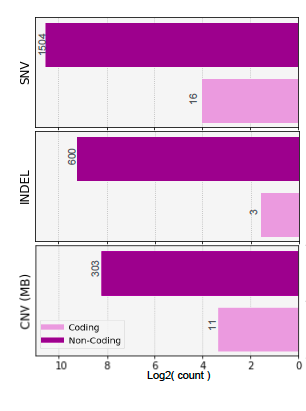
Case Study : 6 y.o. M, Dx of medulloblastoma, Group 4, s/p gross total resection of the cerebral tumor, with spinal leptomeningeal disease. CSF: cytology and ctDNA negative on follow-up.

Representative clinical cases

Overview: The technique of liquid biopsy represents a groundbreaking non-invasive method for tracking cancer progression and remission, particularly during treatment and follow-up phases. Despite its promise, the application of liquid biopsy in pediatric brain cancer via blood samples has faced challenges, mainly due to the minimal tumor presence and scarce genetic mutations in the coding regions. Our research proposes using a whole genome sequencing (WGS)-based, unique mutational signature, derived from a tumor-normal WGS comparison, as a highly sensitive and specific method for identifying mutations in circulating tumor DNA (ctDNA). This approach aims to facilitate effective blood-based monitoring for pediatric brain tumor patients.
Study Approach: To categorize the tumors, we utilized whole genome DNA methylation profiling combined with a machine learning-based classification system. DNA from tumor samples and normal germline DNA from white blood cells were extracted, alongside ctDNA from 1-2 mL of plasma obtained post-operation or during follow-up. We applied WGS for detailed sequencing of DNA from the tumor-normal pairs and plasma samples, achieving 40x coverage for tumor-normal DNA and 20x for ctDNA. Employing the C2i assay, we generated a tailored mutational blueprint for each tumor, complemented by an AI-powered error reduction technique for ctDNA quantification and detection in plasma. This established a patient-centric catalog of somatic mutations, with ctDNA levels assessed during treatment or monitoring intervals, using AI to refine the accuracy by eliminating background noise from cfDNA and enhancing the detection of ctDNA based on the unique mutational signature.
Findings: Our analysis encompassed 7 pediatric brain tumors, including medulloblastomas (Group 3 and Group 4), pediatric glioblastomas (IDH wild-type), an ependymoma (PFA subtype), and a low-grade ganglioglioma. We successfully identified tumor-specific signatures in 5 patients exhibiting clinical disease, with tumor fractions ranging from 0.02 to 0.0005. Conversely, these markers were absent in patients without detectable tumors during blood sample analysis. Notably, in cases of medulloblastoma and glioblastoma, a progressive decline in ctDNA tumor fraction correlated with therapeutic response, as confirmed through imaging.
Implications: The application of a patient-specific WGS-derived tumor signature in ctDNA from blood samples presents a novel and effective method for the sensitive surveillance of pediatric brain tumor conditions. This technique underscores the potential of precision medicine in enhancing the monitoring and management of this vulnerable patient group.
