Introduction
Next-Generation sequencing (NGS) emerged as a revolutionary technology in genomics research aiding precision medicine for almost two decades, but it has its challenges. One of which is the intraspecies contamination which could affect assay’s (1) sensitivity due to the presence of contaminating DNA decreasing the observed allele fraction of variants in the actual specimen (2) accuracy due to the presence of pathogenic variants in contaminating DNA leading to false-positive result.
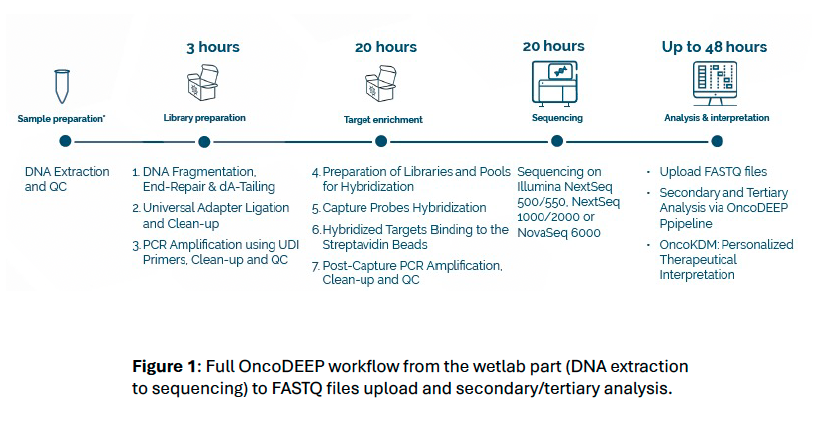
OncoDEEP® Kit is a pan-cancer NGS assay developed with oncology expertise and supported by BioIT solutions. It is based on a comprehensive panel of 638 genes allowing the detection of single nucleotide variant, insertions/deletions, loss of heterozygosity, copy number variation. Additionally, genomic signatures, such as 1p/19q codeletion, microsatellite instability, tumor mutational burden and homologous recombination deficiency can also be assessed.
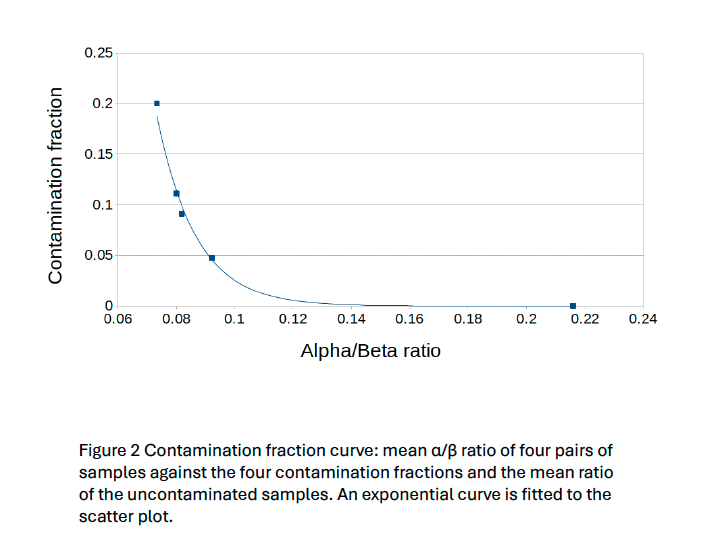
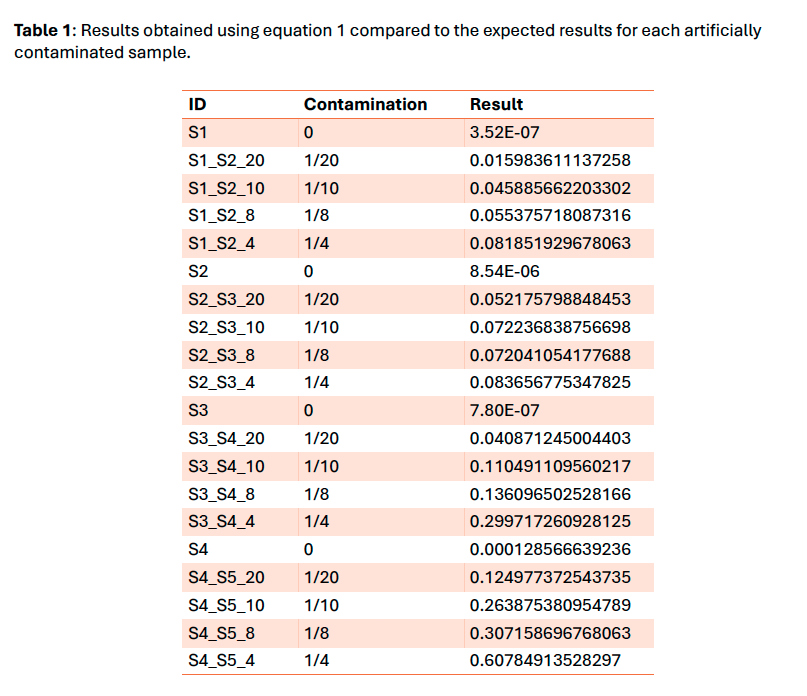
To improve the kit quality control, a check for intraspecies contamination was needed. We selected the methodology presented by Li et al. 2021 in “Contamination Assessment for Cancer Next-Generation Sequencing” due to its ease of implementation, speed and feasibility to scale up for Whole Genome/Exome Sequencing. It is based on an α/β ratio where α is the number of dbSNP variants with 100% of variant allele fraction different from the genomic reference and β is the number of dbSNP variants different from the genomic reference.
Conclusion
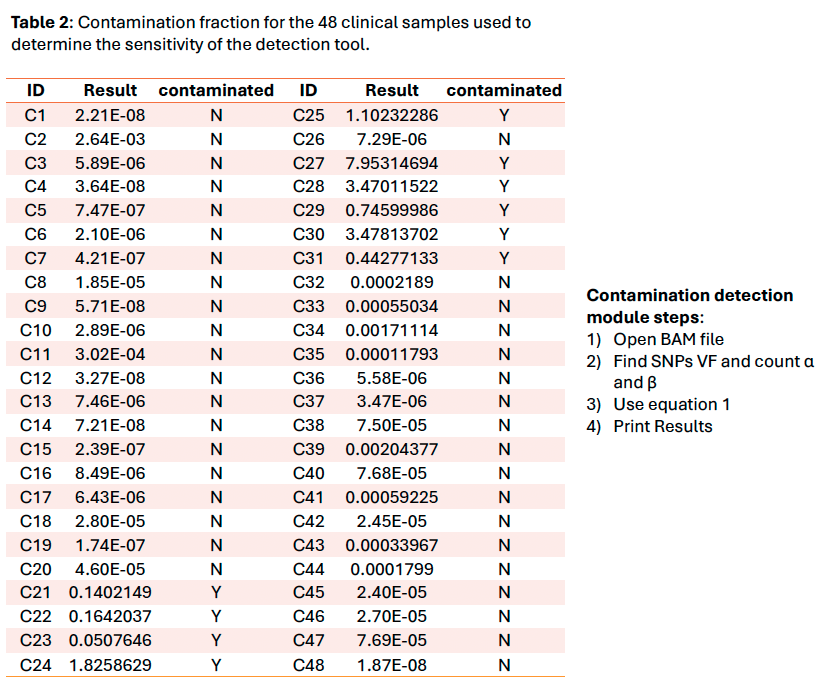
The tool using the Li et al. 2021 method works properly and gives an overview of the contamination level of samples. Nevertheless, this method is only a qualitative tool due to the exponential nature of the formula. The threshold set at 4.5% provided a sensitivity of 100% on the clinical sample’s cohort and is compatible with our variants minimum reporting frequency of 5% for the OncoDEEP kit. The next step would be to verify if it could be used with our liquid biopsy analysis pipeline using Unique Molecular Identifiers which allows a lower minimum reporting frequency.